Targets (outputs & losses)¶
A Target is an output point of the neural network. A Target specifies
how the error must be computed and back-propagated at the output of a layer,
and computes a classification score. A target also specifies how the database
labels must be mapped to the output neurons.
To specify that the back-propagated error must be computed at the output
of a given layer (generally the last layer, or output layer), one must
add a target section named LayerName.Target:
...
[LayerName.Target]
TargetValue=1.0 ; default: 1.0
DefaultValue=0.0 ; default: -1.0
From labels to targets¶
Usually, there are as many output neurons as labels and each label is mapped to a different neuron. This is the default behavior in N2D2: each label in the dataset, by order of appearance when loading the data and label files (files are always loaded in the alphabetical order), is mapped to the next available output neuron. If there is more labels than output neurons, an error is thrown at runtime at the first occurrence of a new label exceeding the output neurons capacity.
This default behavior can be changed using a label-mapping file, where the label mapping is explicitly specified, which is useful to map several labels to the same output neuron for example.
To specify a target in the INI file, one must simply add a LayerName.Target section,
where LayerName is the name of the layer section to which the target must be
associated, as illustrated below:
; Output layer
[seg_output]
Input=...
Type=Conv
KernelWidth=1
KernelHeight=1
NbOutputs=1
Stride=1
ActivationFunction=LogisticWithLoss
WeightsFiller=XavierFiller
ConfigSection=common.config
[seg_output.Target]
LabelsMapping=mapping.dat
The LabelsMapping parameter, when present, is the path to the file
containing the mapping of the dataset labels to the outputs of the layer.
In the present case, there is a single output neuron (per output pixel) in the
seg_output layer, which is a special case where two labels can be mapped
since the activation used is a logistic function. One label can be mapped to
the output value 0 and one label can be mapped to the output value 1.
With more than one output neuron (per output pixel) however, it only makes
sense to map a single label per output.
The label mapping file format is a two-columns, space separated, text table, with the first column corresponding to the name of the label in the dataset and the second column to the index of the associated output neuron.
Two special label names exist:
*corresponding to annotations without valid label (label ID is -1 in N2D2), sometimes referred to as “ignore label” in N2D2;defaultmeaning any valid label name that is not explicitly listed in the label mapping file;
The background name is not a reserved label name, it is simply the name
that is used as DefaultLabel in the Database driver.
Here is an example of label mapping file for the single output layer seg_output:
# background (no defect)
background 0
# padding
* -1
# defect
default 1
Wildcards can be used as well in the name of the label:
* meaning any one or several character(s) starting from this position (non greedy, cannot be used alone as it would refer to the special name for invalid label);
+ meaning any single character at this position.
The actual label mapping of every labels present in the dataset for a given
output target is generated automatically when running the simulation. It is
strongly advised to check this generated file to make sure that there is no
error in the LabelsMapping file and that the mapping is done as intended.
The file is generated in seg_output.Target/labels_mapping.log.dat.
Pixel-wise segmentation¶
Pixel-wise segmentation consists of directly learning a label for each output pixel of the network, typically in a fully convolutional network. Without upscaling, unpooling or deconvolution, the output size is generally smaller than the input size, by a factor S, corresponding to the product of the strides of the successive layers.
In practice, because of the scaling factor S of the network, each output pixel
classify an input macro-pixel of size SxS. It is perfectly possible to get rid
of this scaling factor by rescaling the output to the input size before the
Softmax layer, using bilinear sampling for example. This can be done during
training, in order to precisely back-propagate the pixel-wise annotations, as
the bilinear sampling algorithm is easily differentiable. However, for
inference, the resampling of the output does not add information to the
segmentation (no new information is created when upscaling an image with
context-agnostic algorithms). This is why the scaling factor S may be kept for
inference, without resampling, in order to reduce the computing and
memory cost of processing the outputs.
Other strategies are possible to back-propagate the pixel-wise annotations, which need to take into account the scaling factor S:
Take the majority annotation within the macro-pixel SxS: the label attributed to the output pixel is the label which occurs the most often in the SxS macro-pixel;
Take the majority annotation within the macro-pixel SxS, at the exception of a weak annotation. In this case, any label other than the weak label in the macro-pixel takes precedence over the weak one. In N2D2, this is implemented with the
WeakTargetparameter in[*.target]sections:
WeakTarget=-1means any target other than “ignore” takes precedence. This is useful if the background is ignored. If there is only a few pixels in the macro-pixel that are not background, they take precedence so that the macro-pixel is not considered as background.
WeakTarget=-2means there is no weak label.
Loss functions¶
The loss function in N2D2 is always implicitly defined.
For the Softmax layer or the Logistic activation, the loss is the cross
entropy loss, when used with the WithLoss=1 parameter. Otherwise, the
default loss is the MSE (L2) loss.
The reason is that the error is defined at the output of a layer with the
Cell_Frame[_CUDA]::setOutputTarget() or Cell_Frame[_CUDA]::setOutputTargets(),
which set the value of the input gradient for the cell to (target - output).
These functions are called in the Target class.
So, if a Target is attached to any cell, the corresponding loss
function would be the MSE loss, as the simple difference above is the derivative.
For the softmax or the logistic, the special parameter WithLoss,
when enabled, will simply by-pass the function derivative and directly set
the output gradient of the function to the difference above. This effectively
results to a cross entropy loss with regards to the input gradient of these
functions, as per the mathematical simplification of the cross entropy loss
derivative multiplied by the functions gradient.
Demonstration
The cross entropy loss for a single image is:
Note
M - number of classes (dog, cat, fish)
log - the natural log
y - binary indicator (0 or 1) if class label \(j\) is the correct classification for this image
p - predicted probability that the image is of class \(j\)
The softmax performs the following operation:
To perform the back-propagation, we need to compute the derivative of the loss \(L\) with respect to the inputs \(x_{i}\):
given that \(\sum_k{y_k}=1\), as \(y\) is a vector with only one non-zero element, which is 1.
Target types¶
Target¶
Base Target class.
Base parameters:
Parameter |
Default value |
Description |
|---|---|---|
|
|
Type of |
|
1.0 |
Target value for the target output neuron(s) (for classification) |
|
0.0 |
Default value for the non-target output neuron(s) (for classification) |
|
1 |
The top-N estimated targets per output neuron to save |
|
0.5 |
Threshold for single output (binary classification). |
Labels to targets parameters:
Parameter |
Default value |
Description |
|---|---|---|
|
0 |
If true (1), the data, and not the labels, is the target (for auto-encoders) |
|
Path to the file containing the labels to target mapping |
|
|
0 |
If true (1), labels present in the labels mapping file but that are non-existent in the database are created (with 0 associated stimuli) |
|
-2 |
When attributing a target to an output macropixel, any target other than
|
Masking parameters:
Parameter |
Default value |
Description |
|---|---|---|
|
Name of the |
|
|
-1 |
If >= 0, only estimated targets with ID |
|
0 |
If true (1), the considered estimated targets values are weighted by the estimated
targets values with ID |
Estimated output images parameters:
Parameter |
Default value |
Description |
|---|---|---|
|
-1 |
If >= 0, the corresponding label ID is ignored in the estimated output image |
|
0 |
Hue offset for the first label ID (starting from 0), for the estimated output image |
|
1 |
If true (1), the value in the HSV colorspace is equal to the estimated value. Otherwise, displayed value is 255 regardless of the confidence. |
|
0.0 |
Threshold for estimated value to be considered in the output logs. |
|
jpg |
If left empty, use the database image origin format |
TargetScore¶
The default target, which automatically compute the confusion matrix, confusion metrics and score, for classification or segmentation networks.
Confusion matrix:
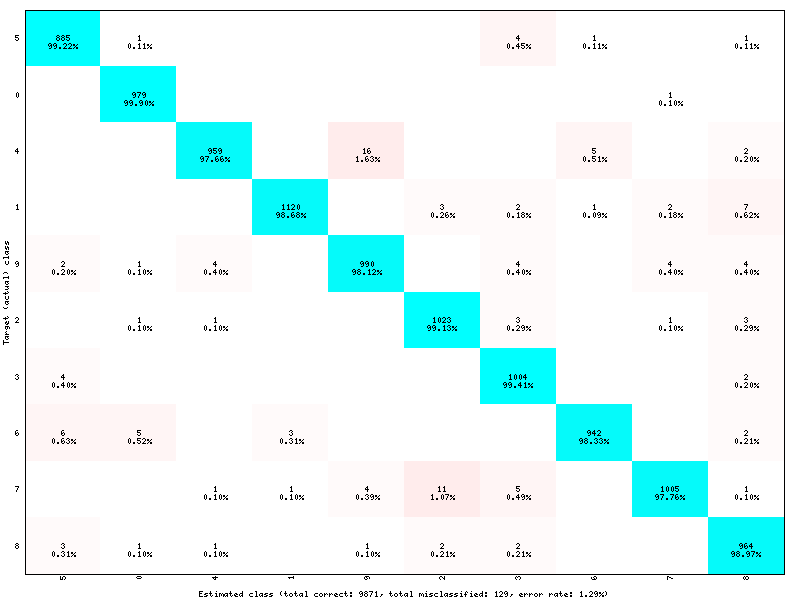
Example of confusion matrix generated by a TargetScore.¶
Confusion metrics:
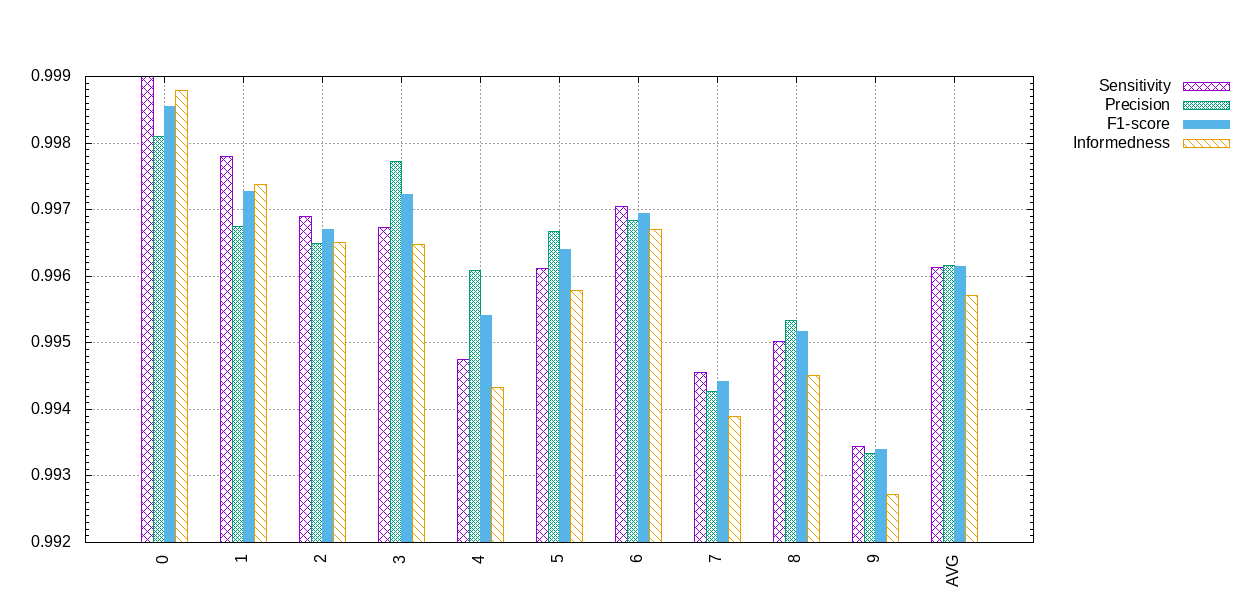
Example of confusion metrics generated by a TargetScore.¶
Score:
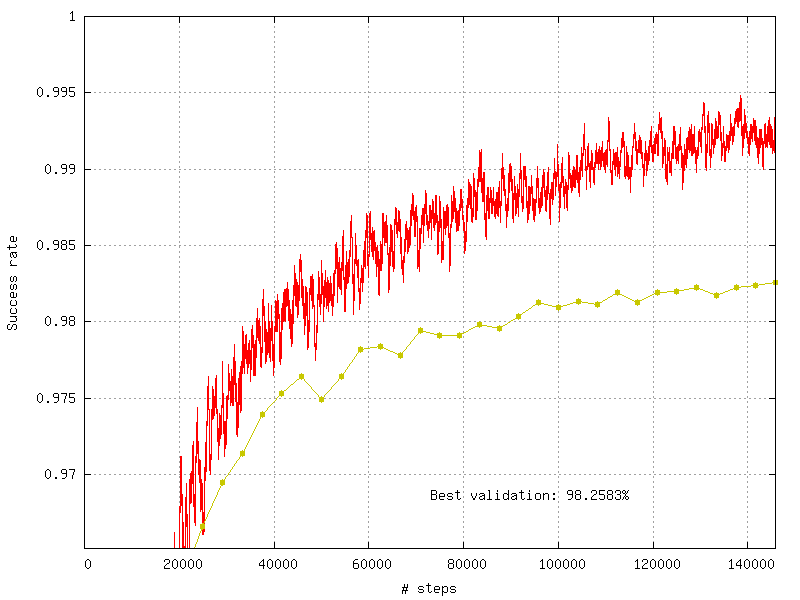
Example of validation score generated by a TargetScore.¶
TargetROIs¶
The TargetROIs allow to perform connected-component labeling (CCL) on
pixel-wise segmentation networks, to retrieve bounding boxes.
This approach is different from classical object detection networks, like SSD or Yolo, where bounding boxes are directly inferred from anchors.
Parameter |
Default value |
Description |
|---|---|---|
|
0 |
Minimum number of macro-pixels above threshold than can constitute a bounding box.
Bounding boxes with fewer than |
|
0.5 |
Minimum overlap (IoU) of a bounding box with an annotation to be considered a match |
|
0 |
Minimum height of the ROI to keep it |
|
0 |
Minimum width of the ROI to keep it |
|
0.0 |
Minimum aspect ratio (width/height) of the ROI to keep it (default is 0.0 = no minimum) |
|
0.0 |
Maximum aspect ratio (width/height) of the ROI to keep it (default is 0.0 = no minimum) |
|
1 |
Maximum horizontal distance for merging (in macro-pixels) |
|
1 |
Maximum vertical distance for merging (in macro-pixels) |
|
1 |
TopN number of class scores to keep for the ROI |